12.4 - Linkage Disequilibrium
It turns out that linkage disequilibrium is a key to doing phasing if we don't have family data. What is linkage disequilibrium?
In diploid organisms (organisms with 2 copies of each chromosome) the parents have two chromosomes each and the offspring gets one from each parent.
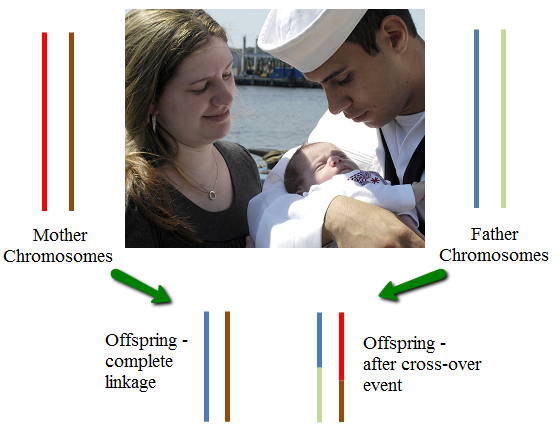
However, processes during meiosis (production of the haploid (one copy of each chromosome) germ cells) crossover events take place in which the two autologous chromosomes exchange material. (Autologous means that these are the "same" chromosomes although there may be differences at the smaller scale.) This is called recombination, because the material in the chromosomes is recombined to form new chromosomes. In many multicellular organisms, the average is 1 - 2 cross-overs in each germ cell. This means that while the offspring obtain half of their genetic material from each parent, most inherited chromosomes are composed of segments arising from both of the chromosomes of the parents. Linkage refers to the probabiity that two segments of DNA are inherited together. If the segments are on different chromosomes, then the linkage probability is 1/4 (because inheritance will be independent, and the probability of selecting one of the two copies from each parent is 1/2). If the segments are on the same chromosome, then the probability depends on the recombination rate between the segments (often called the genetic distance). Although the genetic distance is not the same as the physical distance, thinking of the chromosomes as rigid sticks that get broken and then stuck back together leads to the (accurate) heuristic that segments that are close together have higher linkage than those that are further apart. However, because the chromosomes have weaker and stronger regions, and because cross-over is mediated by biochemical processes, the genetic distance is not identical to the physical distance between the segments.
Two segments are said to be in linkage equilibrium if they are inherited independently. Under random mating without selection you would expect SNPs on different chromosomes to be unlinked. Linkage equilibrium also occurs when the segments are sufficiently distant on the same chromosome. Otherwise, they are said to be in linkage disequilibrium or LD. LD induces association between the genotypes at nearby genetic variants. SNPs close together on the same chromosome are linked.
LD makes it possible and effective to haplotype even when family samples are not included. You can infer what the missing haplotypes are if you have a large sample by using information from the homozygotes to infer the likely pairings for the heterozygotes.

