Lesson 13: ChIP-Seq Data
| Key Learning Goals for this Lesson: |
|
Introduction
The basic paradigm in cell biology is that (almost) all the cells in the organism have the same DNA and that cell types differ due to how they are regulated. Epigenomics is the study of modifications to the DNA that regulate transcription and other cellular functions.
The objectives of the analysis are to locate the locations of interest and determine how the modifications behave under various biological conditions. Because the exact location of epigenomic modications and protein binding sites is not known for most regulatory processes, designing microarray probe sequences is difficult. Thus early studies of protein binding using microarrays had only limited success. Sequencing technology is much better suited to these assays, as the regions of interest can be captured and then sequenced and mapped. Sophisticated assays which target the regions which are protected from DNAse activity by the bound proteins are possible and also use sequencing technology. However, these "footprinting" technologies require sophisticated mapping strategies to identify which proteins are bound at each protected site. In this section we will consider the analysis of one of the simplest assays for epigenomics, which targets regions of the DNA which are bound to specific proteins. Similar statistical analysis methods can be used to assess other assays such as methylation.
A number of epigenetic features are associated with transcriptional regulation including uncoiling of the chromatin and binding of protein complexes to the DNA. ChIP targets the proteins bound to the DNA. There are other assays that target the DNase hypersensitive sites.
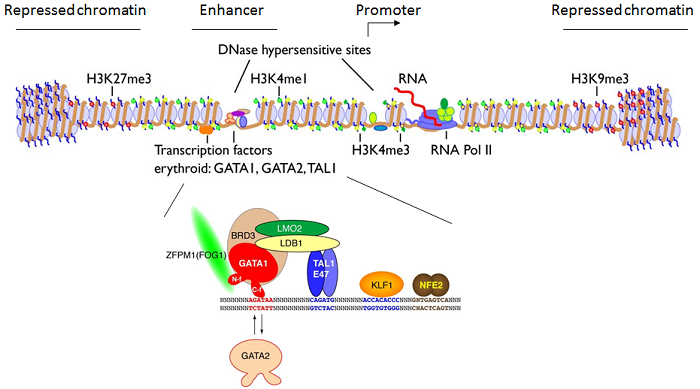
Features captured by ChIP-seq, created R. Hardison
Used with permission.
ChIP-seq can locate the binding sites of specific proteins which may be part of the transcription machinery or may enhance or block the binding of other proteins required for transcription.
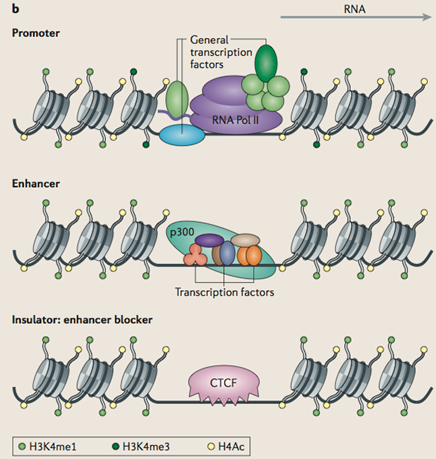
Features of cis-regulatory modules
From Hardison, R. and Taylor, J. (2012) "Genomic approaches towards finding cis-regulatory modules in animals", Nature Reviews, 13: 469.
Used with permission.
General transcription factors are proteins associated with actively transcribed genes. There are also sequence specific transcription factors which bind to regulatory sites adjacent to the gene (cis-regulatory sites) and may enhance or silence transcription. As well, there are proteins that act as insulators, isolating segments of the DNA from activation or silencing.
Epigenetic features play large roles in transcriptional regulation, but understanding these roles requires knowledge of location of the feature relative to the DNA region under regulation. The goal is to determine the locations of these epigenetic features under various conditions. Differential occupancy of sequence specific transcription factors provides insight into gene usage and gene networks.
In the analyses so far, we have been looking at gene expression - the number of transcripts of each gene that can be detected in the tissue. Often, however, we want a more direct measure of what is happening in the cell. ChIP-seq and related analyses such as methyl-seq measure targeted regions of the tissue's DNA by enriching a DNA sample for the targeted regions.
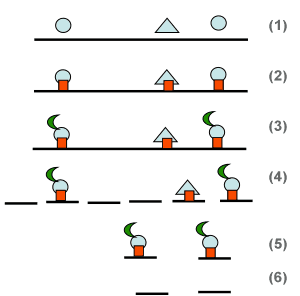
As discussed in Section 1.8 and illustrated in the figure to the right, ChIP-seq targets regions of the DNA (chromatin) that are bound to specific proteins (1). The proteins are stabilized by cross-linking them chemically to the DNA (2). A protein of interest is then tagged with an antibody specific to the protein (3). The DNA is sheered (4), and the fragments bound to the protein are retrieve by chemically retrieving the antibody (immunoprecipitation) (5). Finally, the tag and protein are released from the DNA and washed away, leaving behind the DNA that had been bound (6). This DNA can then be sequenced. The entire process is called ChIP-seq. The objective of the assay is to determine the location of the protein binding sites on the DNA, and whether the protein binds to different sites under different experimental conditions.
Typically the remaining DNA sample is enriched for the bound fragments, but still has a background of miscellaneous DNA fragments. For this reason, we need to do a differential analysis against a sample that was handled similarly except for the antibody binding step. If we are considering several experimental conditions, we may instead do a direct comparison of samples from the different treatments.
As an example, we will be looking at GATA 1 which is a mammalian transcription factor and ask where it attaches to the chromosome and whether it differs between two experimental conditions.

