11.1 - Reference Sets
The feature list comes from our study - it may be the list of differentially expressing features, features with genetic variants, or other feature we have identified. Where do the reference sets come from? There are three main sources of reference sets: search through previous results, pathway diagrams and gene ontologies.
Previous Studies
Sometimes previous studies of the same organism or the same system in a different organism has already indicated a set of interesting features. If it is the same system in a different organism, then your reference set will be homologs of the features in your own study. In a well-studied organism or system there may already be well-curated reference sets. In this situation, typically there is only one reference set and a single test is done.
Pathways and Networks
Over the years, biologists have painstakingly put together biological pathways and networks that show how genes and/or proteins or other cellular processes fit together. These have usually been built manually using data from many sources, although there are now computational tools which can assist in inferring the structure of the network. Network and pathway diagrams may be available from individual papers and/or websites. However, there are also a number of repositories that have collected and annotated the pathways and networks as both diagrams and as machine-readable lists of features involved in the pathway. Most importantly, these repositories impose a uniform annotation on the network which allows the user to search the network and find all the relevant entries.
The network database keeps track of the tags associated with the network diagrams and the list of genes or proteins in the diagrams. These lists are used as reference sets. The reference sets are the genes in the network, or core nodes in the pathway.
The Bioconductor software provides an interface to the most commonly used repositories such as KEGG.
You may be interested in only a single pathway or network, several partitions of a single network, or multiple networks. For example, a search of KEGG for the keyword "serotonin" found 15 networks. If gene set analysis is done using multiple reference sets, multiple testing adjustments should be done.
Gene Onotology (GO)
An ontology is a standardized vocabulary for a field. For example, there is an ontology of medical diagnoses so that anyone reading a medical chart understands the diagnoses.
Once high throughput analyses became available to biology, it quickly became evident that a gene ontology was required to simplify searches and standardize annotation. For example, when annotating the genome, the terms "putative gene, "presumed gene" and "possible gene" were all used to describe sequences for which no transcript or protein had been confirmed, but which resembled coding regions. For genes with actual confirmed transcripts, multiple descriptions are available.
The Gene Ontology Consortium is an organization of scientists who develop the standard terms as well as methodology to annotation genomes using these terms, and tools for retrieving information. Three ontologies are maintained for biological processes (BP), cell components (CC) and molecular functions (MF). As well, important information such how the annotation was determined is retained with the annotation. For example, some terms are applied to a gene due to experimental results when the gene is up or down-regulated; other terms are applied due to sequence homology with a gene in another species with known function in the other species.
Many species are represented in the Gene Ontology database. The ontology for an organism is constructed by an organism consortium. When a new genome comes out, for instance, interested scientists may volunteer to create an ontology based on existing ontologies, and then assigning the terms to the features of the new genome.
The ontology's are organized as graphs. Each node is a term. The leaf nodes are more specific instances of the term. For example, a term might be "response to stress" and the leaf nodes might be "response to DNA damage" and "response to heat shock". However, a leaf node might have multiple parents, so the graph is not necessarily a tree. Here's an example of a Go graph. It is only a small piece of the biological process:
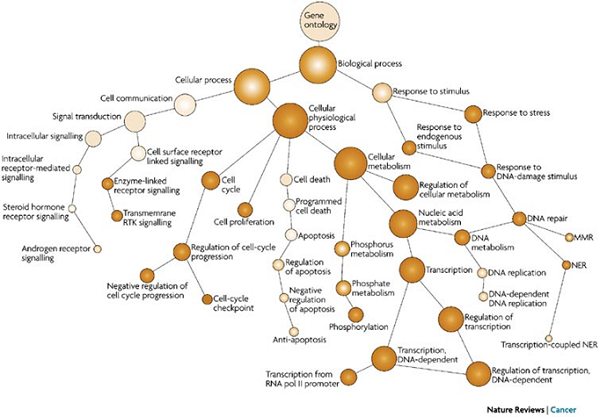
(from P. Hu, Computational prediction of cancer-gene function. Nature Reviews Cancer 7, 23-34 (January 2007), used with permission.)
The nodes of the graph are not independent. Any gene assigned to a node in the graph is also assigned to its parent nodes. Each node is associated with a set of genes that have this term in their annotation. The gene set at any node is a subset of the gene set at any parent node.
There is also reduced ontology that people use to produce pie charts and bar charts that are often seen in papers. This is called GOSlim. GOSlim cuts the tree at one level, so that each gene is assigned to a unique node.
There are two main ways to use the information in GO for gene set analysis.
Firstly, you can download a small number of categories, usually with GOSlim and then test those categories. Usually you would select categories so that do not overlap too much, so that you can assume the tests are independent. Ordinarily a chi-squared test is performed in each category but you should include multiple testing adjustments.
Another mode of using GO for gene set analysis is to select one of the 3 ontologies and test at every node. There are several software packages available for this - we will use one of the Bioconductor packages. Since the reference set at each node is nested within the set defined by the parent node, this leads to multiple correlated tests. Multiple testing is complicated due to lack of independence among the tests. The software we will use works in two modes - the unconditional mode reports the significant tests at every node. The conditional mode reports only the child node if the statistical significance of the parent can be attributed to enrichment or depletion of the child.

