9.3.1 - R Scripts
1) Acquire Data
Diabetes data
The diabetes data set is taken from the UCI machine learning database repository at: https://archive.ics.uci.edu/ml/datasets/Pima+Indians+Diabetes.
- 768 samples in the dataset
- 8 quantitative variables
- 2 classes; with or without signs of diabetes
Load data into R as follows:
# set the working directory
setwd("C:/STAT 897D data mining")
# comma delimited data and no header for each variable
RawData <- read.table("diabetes.data",sep = ",",header=FALSE)
In RawData, the response variable is its last column; and the remaining columns are the predictor variables.
responseY <- as.matrix(RawData[,dim(RawData)[2]])
predictorX <- as.matrix(RawData[,1:(dim(RawData)[2]-1)])
For the convenience of visualization, we take the first two principle components as the new feature variables and conduct k-NN only on these two dimensional data.
pca <- princomp(predictorX, cor=T) # principal components analysis using correlation matrix
pc.comp <- pca$scores
pc.comp1 <- -1*pc.comp[,1]
pc.comp2 <- -1*pc.comp[,2]
2) K-Nearest-Neighbor
In R, knn performs KNN and it is in the class library. Again, we take the first two PCA components as the predictor variables.
library(class)
X.train <- cbind(pc.comp1, pc.comp2)
model.knn <- knn(train=X.train, test=X.train, cl=responseY, k=19, prob=T)
In the above example, we set k = 19 due to the follow reason. To assess the classification accuracy, randomly split cross validation is used to compute the error rate. The samples are split into 20% for test dataset and 80% for training dataset. Fig. 1 shows the misclassification rates based on different k in KNN.
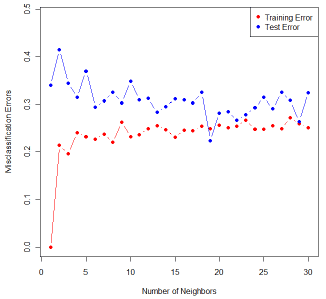
Figure 1: Training and test error rates based on cross-validation by K-NN with different k’s.
Based on Fig. 1, k = 19 yields the smallest test error rates. Therefore, we choose 19 as the best number of neighbors for KNN in this example. The classification result based on k = 19 is shown in the scatter plot of Fig. 2. The red circles represent Class 1, with diabetes, and the blue circles Class 0, non diabetes.
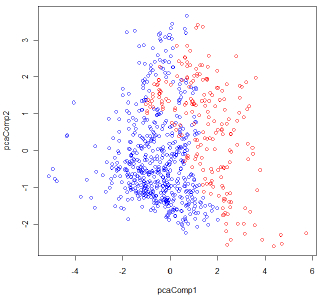
Figure 2: Classification result based on KNN with k = 19 for the Diabetes data.
The knn function also allows leave-one-out cross-validation, which in this case suggests k=17 is optimal. Results are very similar to those for k=19.
After completing the reading for this lesson, please finish the Quiz and R Lab on ANGEL (check the course schedule for due dates).

