1.2 - What Can We Measure?
These days, given enough time and money, we can measure just about anything about nucleic acids: their sequence, their abundance, what is binding to them and where, how tightly they are coiled, how they interact with cell components, and so on.
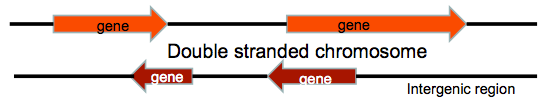
When measuring DNA, we might measure both strands simultaneously, or each of the strands singly. Although the sequence information is redundant due to complementarity, coding is done from the single strands, and so coding information is strand specific. The two strands of the chromosome are polar, so they have a coding direction. Some genes are on one strand and some genes are on the other, and they can overlap. The genes on sister strands will code in opposite directions. Between the genes is an area called the intergenic region. Originally it was thought that the intergenic regions were "junk" but increasingly it has been found that these regions may encode small functional units or be involved in gene expression regulation. Biologists often measure some of the small functional RNAs, such as silencing RNAs and are also interested in regulatory regions.
Transcription is the process of going from the DNA (the storage molecule) to RNA (the biologically active molecule). The region of the chromosome upstream (before the start) of the genes is called the promoter region. Proteins called transcription factors bind to this region and get transcription started. Transcription factor binding sites (and other protein binding sites) are often of interest. The set of all transcripts is called the transcriptome. Gene expression analysis usually involves measurement of the transcriptome or the protein-coding part of the transcriptome.
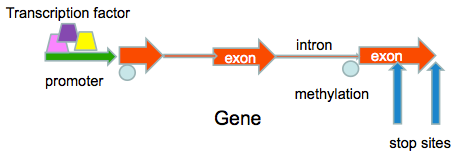
In eukaryotes, many genes are made up of short contiguous chunks on the chromosome. The exons are the parts that have the protein encoding bases (the codons) and in between these are spacers called introns that have regulatory mechanisms. The regulatory mechanisms determine which exons are used to create transcripts. The gene also has start and stop sites to direct where transcription begins and where it ends. The exons can be put together in various ways to create different proteins, often called isoforms. One way to think of the exons is like syllables (in English) or like characters (in Chinese). They can be combined to make many words.
The set of all exons is call the exome. The total bases in the exome is usually only a few percent of the total bases in the DNA. Recent technology has enabled biologists to sequence just the exome, particularly when looking for genomic variants. However, this may miss important parts of the genome both because exons may not be recognized and because the regulatory regions are excluded.
An important type of gene regulation is called methylation. It is the addition of a methyl group to what is called a CpG site, a location on the chromosome where C and G nucleotides are adjacent. Methylation is what is called an epigenetic factor - it is a direct but reversible chemical change in the DNA. Various environmental and evolutionary processes can create methylation, and some of this methylation is passed on to offspring. There are a number of interesting biological processes that seem to be regulated by methylation such as stress reactions. Smoking, drinking and other drug-like substances can change methylation patterns. The location of methylation sites and their methylation state is another genomic feature of interest to biologists.

