1.9 - Microarrays
A microarray is a substrate to which are attached millions of single-stranded (c)DNA complementary to the (c)DNA you wish to detect. They come in two flavors, either the substrate is a small plastic or glass slide like a microscope slide or a plastic bead. There can be from a few thousand to 1 million probes each consisting of thousands of single strands with the same sequence. The probes are attached to the substrate.

A labeled sample of DNA or cDNA is allowed to hybridize or attach to the probes. Each fragment should bind only to a complementary probe. After hybridization, the substrate is washed which removes any material that didn't bind to a probe. Each probe on the microarray should be bound to a sample of the targeted complementary DNA. The quantity of bound (c)DNA is expected to be roughly proportional to the amount of that type of (c)DNA in the labeled sample.
A dye intensity for each probe is summarized by a scanning microscope which essentially takes a photo of the microarray at the wavelength of the label. The raw data is the intensity of reflectance of the label at each pixel. This intensity is expected to be proportional to the amount of material in the labeled sample.
The most fundamental data is a digitized photo of the array giving the label intensity.
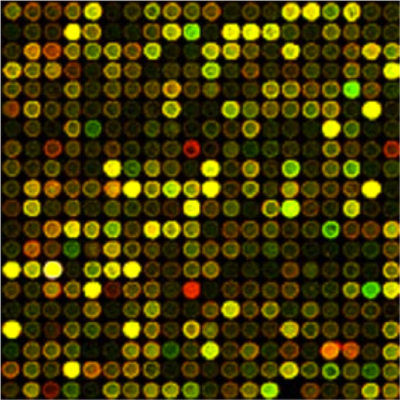
This is a digitized and color-enhanced photo of an older array. You can tell that it is older because the spots on this image are not uniform in intensity due to the technology used to create the probes. On the newer microarrays used since 2005 the probes are printed on the array surface using print technology and appear absolutely perfect at this scale.
Microarrays come in many formats. The microarray pictured is sometimes called a "spotted" microarray, due to the original printing technology. Each spot represents a cluster of probes of the same oligonucleotide (fragment of DNA). Usually the entire cluster is referred to as a probe. Either one sample (single channel) or two samples with different labels (two channels) are hybridized to this type of microarray. There may be multiple probes for a single gene, sometimes actual duplicates and sometimes different oligonucleotide sequences (oligos) from the same gene. Often several identical microarrays are printed on a single glass or plastic slide, with a barrier around each to keep the samples from mingling. When an entire experiment can be run on a single slide, uniformity of the hybridization conditions is assured.
Another older format that is still in use is the Affymetrix microarray. Affymetrix was the first manufacturer to synthesize the probes on the array surface, allowing very accurate probe synthesis and close spacing. Each gene is represented by one or more "probe sets". A probe set is a collection of probes made up of 25-mer oligos selected from the DNA sequence. The older arrays had both "perfect match" probes, which exactly match the reference genome (at least the reference when the array was first designed) and "mismatch probes" which are paired with the "perfect match" probes but differ in the central nucleotide. The "mismatch probes" were supposed to assist with background correction and correcting for cross-hybridization; however as evidence mounted that this strategy did not work well, newer Affymetrix microarrays have only "perfect match" probes. Usually a gene is represented by 9 to 11 "perfect match" probes, with the number of probes per probe set constant in any given microarray. Probes in the probe set are chosen for uniformity of hybridization and uniqueness. On many microarrays, most of the probes are selected from the 3' end of the gene. Cartoons showing the construction of a probeset can be found at the Affymetrix website. Affymetrix microarrays are still very popular, especially for genotyping using a set of known SNPs. This is because the "prefect match"/"mismatch" pair is readily replaced by the 2 SNP variants at a locus.
Another microarray technology is the bead array. We will not be discussing bead arrays in this course, but you can read about them at https://www.ncbi.nlm.nih.gov/probe/docs/techbeadarray/.
The analysis technology for microarrays, from probe intensity summary of the image pixels through the analysis of gene expression or gene variant is very mature. Different preprocessing methods are required for each type of array to achieve a summary. For gene expression microarrays, this might include probe location detection, summary of the pixels in the area defining the probe, background correction, assembling the probe intensities into gene intensities.
Here is an example of what the data might look like for a spotted two-channel array. This is an intermediate probe summary produced by the scanner.
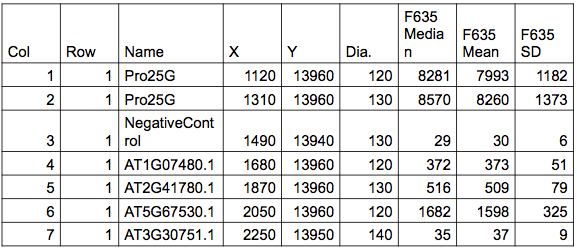
The table is a rectangular array. It has row and column numbers, probe name, X and Y coordinates for the center of each probe and the diameter of the probe. Then the intensities are reported for each probe. Each pixel of the photo represents an intensity. Since the probes consist of many pixels, a summary such as the median, mean and standard deviation are given so that you have choices to use for analysis. On modern microarrays the median and the mean are practically the same because the spots are accurate and uniform. However, if you use historical data, they might be quite different. There are also summaries of the background local to each probe.
The probes are designed to detect various features by selecting parts of the reference genome or transcriptome to match. For this reason, microarrays are quite flexible. You can use them for gene or exon expression, SNP detection, ChIP, methylation and other features of interest.
Microarrays have some pros and cons. There are an older technology so we know in great detail how to process microarrays, both in the lab and statistically. They are relatively cheap. Data storage is minimal - you can store the outcome of your entire experiment on your keychain flash drive.
However, microarrays are species or genotype specific, although the same array can sometimes be used for closely related species e.g. humans and chimpanzees. A serious downside is that you have to know to advance what you're looking for because microarrays require known sequences be used as probes. You can't capture anything for which you don't have a probe. In addition, genetic variation affects how intensely the complemental DNA hybridizes.
Sequencing technologies by contrast can detect unknown sequences. However, you will not know what you have found unless you have a reference sequence or have enough sequences to create a de novo reference. This sounds contradictory, but is not. For example, since we have sequenced almost all of the human genome, previously unknown protein binding sites, methylation sites, etc can be identified by sequencing and their locations on the genome identified by matching to the reference.
A strategy that is now commonly used for species with few genomic resources is to do a small amount of transcriptome sequencing and use it to create microarray probes. Annotation can be challenging, but once highly expressed or differentially expressed probes are identified, lower throughput technologies can be used to identify the genes.
As well as DNA microarrays, there are now microarray technologies for proteins. Instead of DNA probes, the microarray is printed with "capture probes" which bind to the proteins of interest. The probe intensity is then measured, yielding a set of intensities that are similar to DNA microarray data. While preprocessing of the arrays may differ from DNA arrays, the statistical analysis of the processed data is quite similar.

